Understanding how the diversity of life on earth arose and is maintained is the foundational question in evolutionary biology. Our lab studies the process of speciation from a genetic perspective. We are particularly interested in what can be learned about the process of speciation from cases where hybridization, or mating between distinct species, occurs. Although hybridization between species is common, many hybridizing species remain genetically, ecologically, and behaviorally distinct. Understanding why this is so from the molecular to the population level is at the core of research in our lab.
What is the genetic architecture of reproductive isolation between hybridizing species?
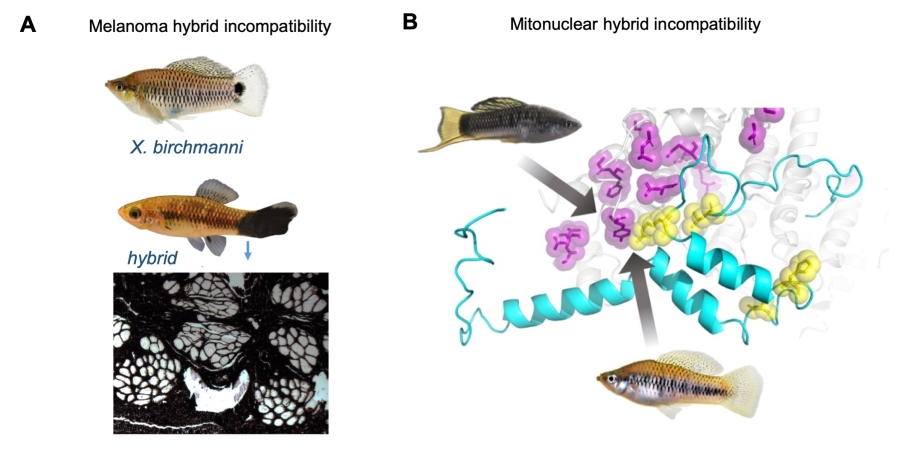
Biologists have long appreciated the fact that when species hybridize, hybrids often suffer reduced viability or fertility, presumably due to genes that have functionally diverged between the parental species and no longer interact properly in hybrids. Despite a century of work documenting this phenomenon, we know very little about which genetic interactions cause hybrid incompatibilities and the evolutionary and molecular paths through which they arise. This is especially true for species that naturally hybridize, many of which have historically been difficult to study in the lab. We use naturally hybridizing species of swordtail fish as a powerful model system to study the genetic basis of hybrid incompatibilities and uncover their evolutionary history. Current work in the lab is focused on the role of coevolution in regulatory networks and protein complexes in driving hybrid incompatibilities.
How does selection shape introgression in the genome?

Many species have hybrid genomes – that is, large proportions of their genomes are derived from introgression from other species. In some human populations, more than 2% of the genome is derived from admixture, while in some swordtail fish species, as much as 30% of the genome is derived from hybridization. Yet except for a few cases, we understand little about how the genome evolves and stabilizes after hybridization, and what the functional consequences of introgressed regions are. This puzzle is one of the major focuses of the lab. To understand this process, we study both ancient and contemporary hybridization events, and complement this with studies of lab generated hybrids and modeling approaches.
Another open question is how many regions of the genome contribute to hybrid breakdown, what their distribution in the genome is, as well as how strongly selection acts upon them in hybrids. As part of our work addressing this question, we are also interested in understanding other causes of selection on hybrids, such as hybridization load, and developing models and methods that can help us better understand mechanisms of selection on hybrids.
What is the genetic basis of adaptive differences between species and how do they impact hybridization?
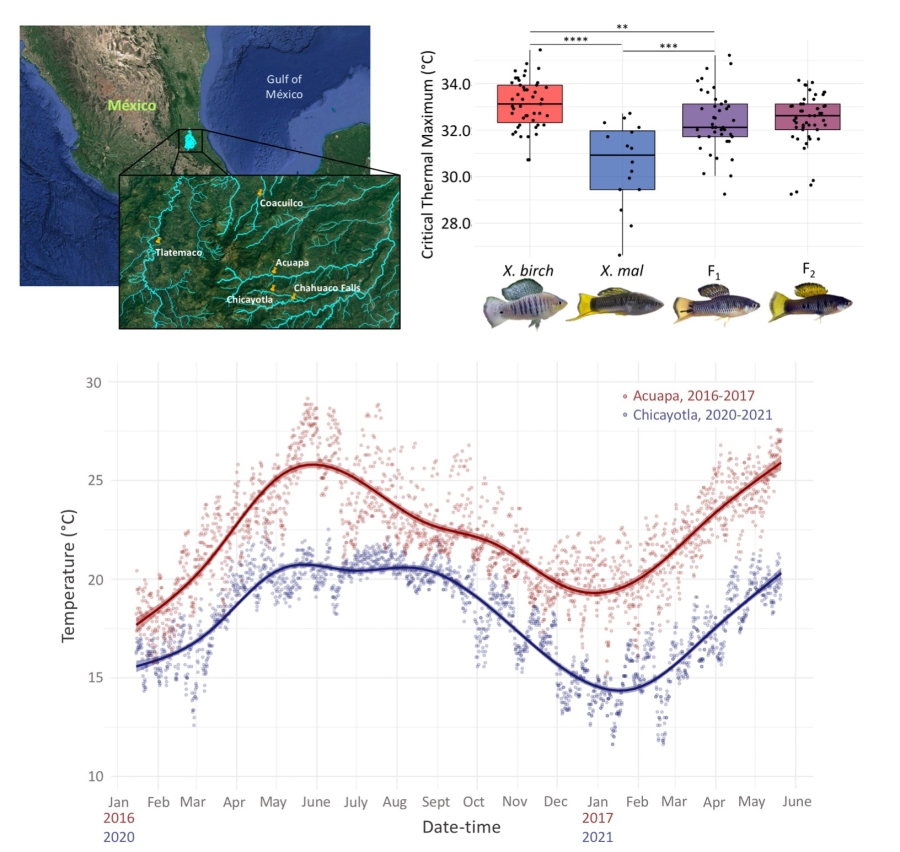
Hybrid incompatibilities are just one of many barriers that arise as species diverge. Adaptation to the ecological environment and coevolution in sexually selected signals and preferences also drive phenotypic and genetic divergence between species. Our lab is interested in characterizing the genetic basis of such differences between species and uncovering how these loci impact the movement of genes between species. Current project are focused on the genetic basis of adaptation to different thermal environments and the drivers of sexually dimorphic traits within and between species.
How do recombination rate and mechanism shape introgression?
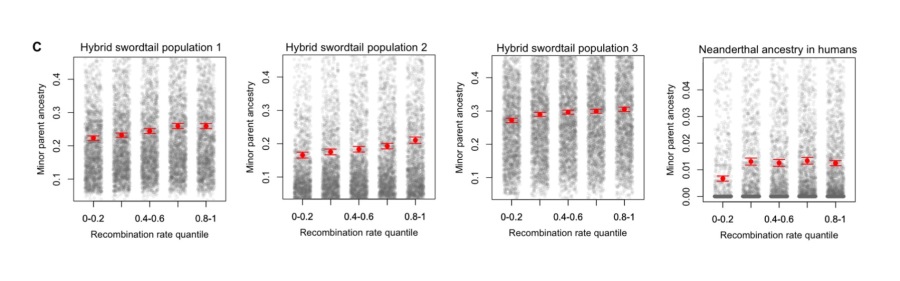
While it is increasingly clear that many species hybridize, we know comparatively little about the ecological, genetic, and evolutionary forces that shape hybrid ancestry in the genome. Previous work in our group has suggested that local recombination rate is a key parameter in this process, with ancestry from the minor parent species being preferentially retained in regions of the genome with the highest recombination rates. This is likely because in regions of high recombination, haplotypes derived from the minor parent can more quickly become uncoupled from linked sites that are under selection in hybrids. Understanding how commonly interactions between recombination and selection on hybrids generates these patterns in the genome is a current focus of the lab. Further, there is remarkable diversity in how different species regulate recombination and as a result where in the genome recombination occurs, which have implications for dynamics of introgression between species.
How do sexually selected traits influence hybridization and the evolution of hybrid populations?

Many species exhibit conspecific mate preferences, but for hybridization to occur between species these preferences must break down. As a result, fragile or context-specific mate preferences are likely to be a major mechanism mediating hybridization between species. Further, in hybrid populations combinations of sexually selected traits and preferences from both parental species could lead to assortative mating in hybrids, different rates of gene flow between hybrids and their parental populations, or even reproductive isolation between hybrid populations and their parental species. Exploring the impacts of sexual selection on hybridization and evolution of hybrid populations is a new focus in the lab.
CICHAZ field station – Calnali, Hidalgo, Mexico
We work with a fantastic field station in the state of Hidalgo, Mexico. For more information about working at CICHAZ, see the CICHAZ homepage



